Twenty years ago, when researchers from the Human Genome Project announced that they had completed the first-ever sequence of an entire set of human DNA, the discovery was heralded as comparable to “splitting the atom or going to the moon.” The announcement forever transformed the fields of genomics, biology, and medicine, allowing researchers to investigate more complex questions and seek more complicated answers related to the human body and how to treat disease.
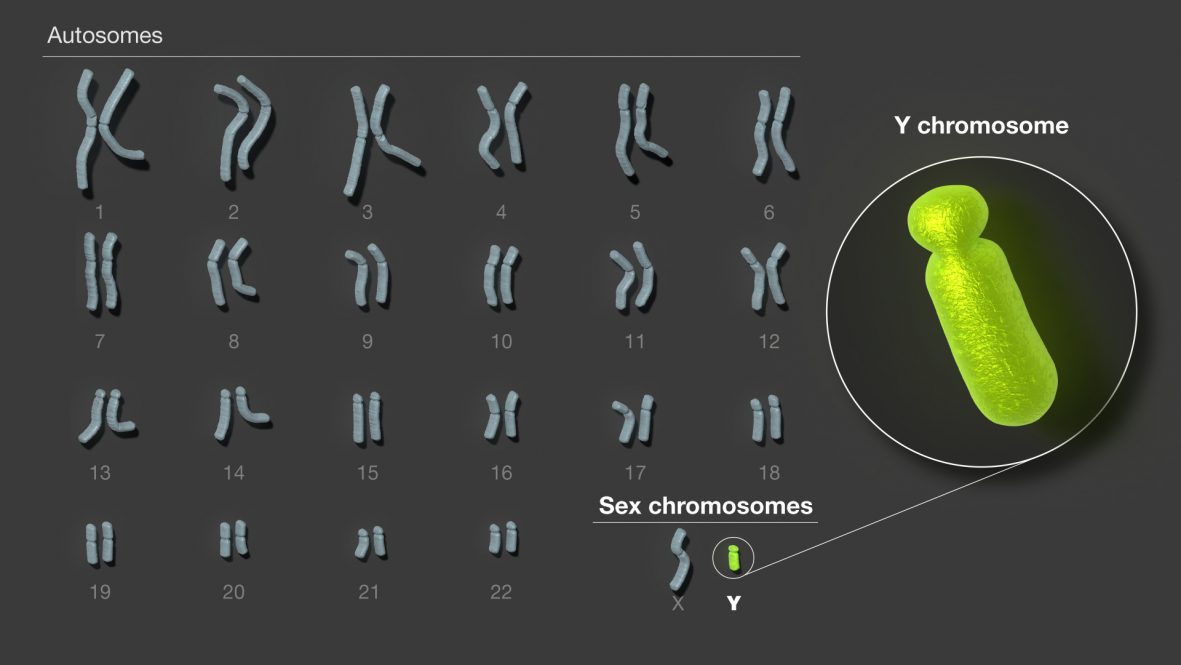
But the researchers had not, truly, completed a full human genome sequence. Their landmark findings were defined as “highly accurate” and “highly contiguous,” with “the only remaining gaps corresponding to regions whose sequence cannot be reliably resolved with current technology.” A truly completed sequence was published last year by the Telomere-to-Telomere (T2T) consortium, an international scientific collaboration featuring the work of geneticist Rachel O’Neill, director of UConn’s Institute for Systems Genomics (ISG) and Board of Trustees Distinguished Professor in the Department of Molecular and Cell Biology, and her lab.

The T2T consortium sequenced the entire human genome from a cell with XX chromosomes. The findings were still groundbreaking, but since approximately half of the human population has XY chromosomes rather than XX, researchers knew that their work wasn’t done.
“There are a lot of clinical outcomes linked to the Y chromosome,” notes O’Neill.
In two new papers, published August 23, 2023, in the journal Nature, O’Neill and her collaborators offer an in-depth analysis of the Y chromosome for the first time, augmenting the findings of the T2T consortium.
The first paper, a publication of the T2T consortium, is the first complete Y chromosome assembly from an individual (T2T-Y), The second, a collaboration with The Jackson Laboratory for Genomic Medicine (JAX), presents the Y chromosome sequences from 43 unrelated individuals, providing a foundation for future inquiry into how genetic diversity within the Y chromosome impacts health outcomes.
O’Neill’s lab worked with several other key investigators and labs to accomplish this monumental feat. In particular, she notes, the lab team of Adam Phillippy at the National Institutes of Health provided the assembly work for the chromosome and pioneered the work.
“We did all of the repeat annotations,” she says, “which were pretty extensive, because more than 50% of that chromosome is repetitive.”
The work benefited from the advancement of sequencing technology, which has come a long way since the Human Genome Project’s first sequencing attempt completed in 2003. Long-read sequencing technology, O’Neill explains, is key for successfully interpreting sections of DNA that are highly repetitive.
Without the long-read tech, O’Neill says, “it was like trying to put a book together when all you have is a pile of words.” The new technology, in contrast, “gives you whole sentences, paragraphs, pages.”
As the first paper, led by Arang Rhie of Phillippy’s group, was entering the publication process in Nature, O’Neill was contacted by researchers at JAX, who were also in the process of Y chromosome assembly. JAX professor Charles Lee and associate research scientist Pille Hallast were heading up a team that analyzed and sequenced 43 different Y chromosomes from distinct individuals, with nearly half coming from African descent (significant because the T2T-Y chromosome sequence had been accomplished using DNA from an individual of European descent).
Once again, O’Neill’s lab assisted with the repeat annotations, using long-read technology to determine the specific number and order of repeating sections on the chromosome. Repeating sections of DNA – which may differ in number or sequence between individuals – account for most of human genetic variation.
The T2T-Y chromosome sequence identified an additional 42 protein-coding genes that had not previously been studied and added an additional 30 million base pairs to the existing reference human genome, while the JAX sequencing work contributed 43 new unique Y chromosome sequences.
“Taken together, these two papers provide intriguing insights into human Y chromosomes, reveal the highly variable nature of Y chromosomes across individuals, and provide an important foundation for future studies on how they may be contributing to certain disorders and diseases,” notes the JAX release.
They represent a monumental advance in the field of genomics and another exciting publication for the lab of O’Neill, who continues to pioneer work exploring the effects of repetitive elements in the human genome.
This work was supported in part by a U24 grant from the National Human Genome Research Institute (#HG007497).